Animal Gut Microbiome Testing Service
16S rDNA-BASED MICROBIAL DIVERSITY ANALYSIS
One of our Next Generation Sequencing Services...
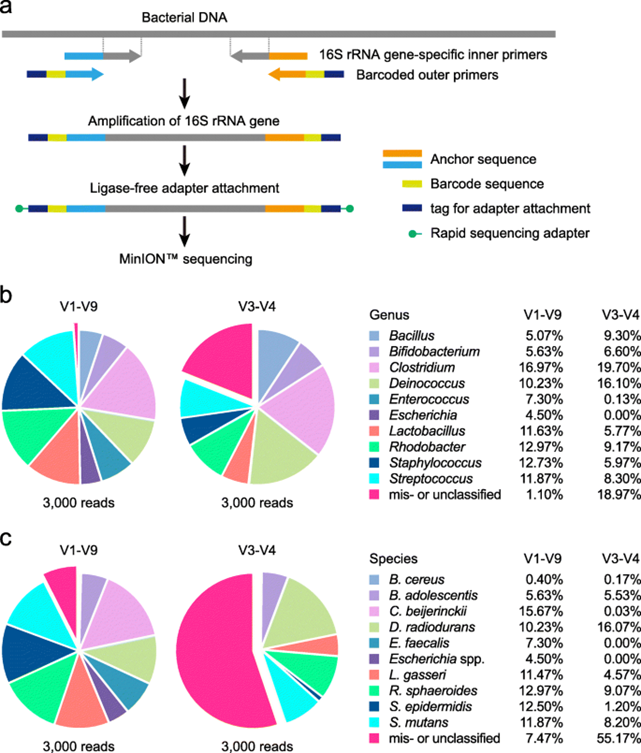
Microbiome Sequencing
Animal Gut Microbiome Testing is a process of analyzing the genetic material of microorganisms present in a given biological sample, such as soil, water, gut, or skin, using high-throughput DNA sequencing technology.
The microbiome NGS service provides a comprehensive analysis of the microbial community composition, diversity, and function present in a particular environment, which can help in understanding the role of microbes in various biological processes.
Animal Gut Microbiome Testing Process
1. Sample collection (by the customer): The first step is to collect the biological sample that contains microbial communities. The sample collection process varies depending on the type of sample, and it is crucial to ensure that the sample is representative of the microbial community in the environment.
2. DNA extraction: The next step is to extract the DNA from the collected sample using various DNA extraction protocols. The quality and quantity of the extracted DNA determine the success of downstream analysis.
3. Library preparation: The extracted DNA is then converted into a library of DNA fragments, which are amplified and sequenced using NGS technology. This step involves adding adapters to the DNA fragments and PCR amplification to generate sufficient material for sequencing.
4. Sequencing: The libraries of DNA fragments are then sequenced using NGS technology, which generates millions of short DNA reads. The sequence data generated in this step can be used to determine the composition, diversity, and function of the microbial community present in the sample.
5. Bioinformatics analysis: The final step is to analyze the sequence data using various bioinformatics tools to identify the microbial taxa and their functions present in the sample. This step involves quality control, filtering, assembly, and annotation of the sequence data.
Microbiome
16S Amplicon-based Microbial Profiling Specifications
Sample
≥20 µl, ≥5 ng/µl High quality DNA
Library preparation
KAPA HiFi HotStart Ready Mix Kit (Roche) and Nextera XT v2 Index Kit (Illumina)
Inclusions
- Sequencing
- Bioinformatics
- Raw data transfer
Microbial diversity analysis includes sample quality control, library preparation, final QC, sequencing and full bioinformatic analysis (filtering, trimming, and metagenomic report*); free data transfer of FastQ files and Reports
Sequencing metrics
Performed on Illumina MiSeq/NextSeq 2000 with 2×300 bp paired-end run; Seq throughput: 0.2M PE read per sample; Q30 ≥ 70%
Quality control
Full quality assurance and guaranteed specifications
Turnaround time
min. 2 – max. 6 weeks


